Innovation to Impact. See you at the July ESIP Meeting.
A spatiotemporal indexing approach for efficient processing of big array-based climate data with MapReduce
Climate observations and model simulations are producing vast amounts of array-based spatiotemporal data. Efficient processing of these data is essential for assessing global challenges such as climate change, natural disasters, and diseases. This is challenging not only because of the large data volume, but also because of the intrinsic high-dimensional nature of geoscience data. To tackle this challenge, we propose a spatiotemporal indexing approach to efficiently manage and process big climate data with MapReduce in a highly scalable environment. Using this approach, big climate data are directly stored in a Hadoop Distributed File System in its original, native file format. A spatiotemporal index is built to bridge the logical array-based data model and the physical data layout, which enables fast data retrieval when performing spatiotemporal queries. Based on the index, a data-partitioning algorithm is applied to enable MapReduce to achieve high data locality, as well as balancing the workload.
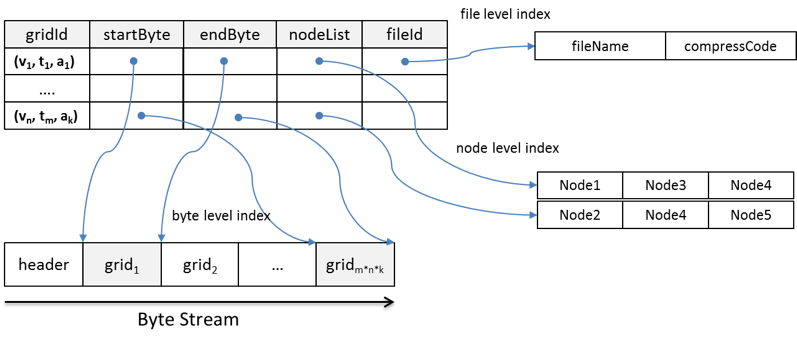
The proposed index structure is illustrated in Figure 1. The index contains five components: gridId, startByte, endByte, nodeList, and fileId for three levels of the index: byte level, file level, and node level. To build the index, the values of the five components are extracted from the array-based data stored in HDFS using an customized access library (e.g. NetCDF for Java).
-
gridId is the bridge between the logical data view and physical data layout. It consists of three parameters in the logical view: variable, time, and altitude. startByte and endByte are the byte-level indices that record the exact byte location of the grid in a file. fileId is the file-level index that records which file a grid belongs to and how data in that file is compressed. The byte- and file-level indices enable each grid to be read directly from a file's byte stream using the file system's native I/O method. This improves efficiency by eliminating the need to consult metadata to retrieve a piece of data from a large file.
-
nodeList records the node location where grids are physically stored. The number of nodes in each list is equal to the replication factor. Some grids are split into two blocks. For these grids, a node in the nodeList may only store part of the grid. However, since the block size is generally much larger than the grid size, most grids remain intact within blocks and nodes across the HDFS. An effective grid assignment strategy allows most grids to be read locally, which maximizes data locality, an important factor affecting performance in MapReduce.
Figure 1. Structure of the spatiotemporal index
HDFS partitions large files into many logical splits, and then assigns these splits to physical data blocks on physical nodes. How these splits are partitioned and assigned directly impacts data locality, which has a dramatic affect on the performance of MapReduce. Therefore, a partition strategy is developed to uses the spatiotemporal index to optimize processing performance by 1) keeping high data locality for each map task, 2) balancing the workload across cluster nodes, and 3) generating a proper number of map tasks to minimize the overhead.The partition strategy is composed of two parts: grid assignment and grid combination. For the grid assignment, it assigns each node with similar number of grids to keep the workload balanced between the nodes. It also considers the physical locations of the grids to enable grids read locally. For the grid combination, it organizes a similar number of grids as an input split, and deliver them to each map task. Then each map task will process the nearly equal volume of data. Figure 2 shows the overall process for index-based parallelization with MapReduce
Figure 2. Index-based parallelization with MapReduce
If you are interested, please click this link to find the detailed paper.